
Bacterial Genomics
Unlock the secrets of microbial genomes to understand the evolution of stress resistance.
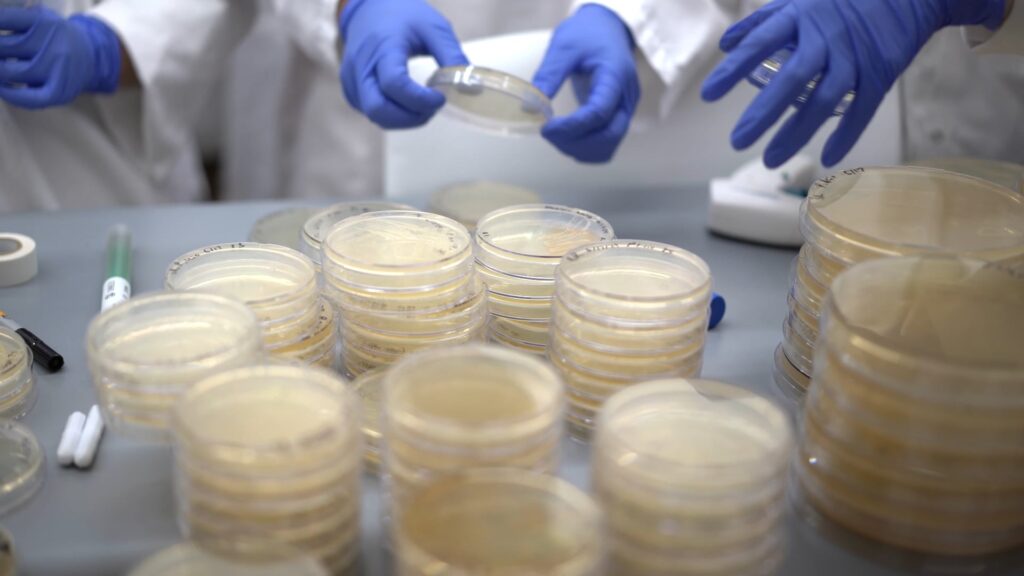
About the Program
Participants in the Summer Science Program in Bacterial Genomics study the evolution of stress resistance in a non-pathogenic bacterium. By introducing ever increasing amounts of stressors, the population is forced to evolve or perish – and we can study that evolution by analyzing their DNA and how it changed from the ancestral strain. Through this process, participants gain hands-on experience with the microbiology, molecular biology, genetics, genomics, and bioinformatics that they’ll need to go from the output’s seemingly random ATGCs to annotated genomes and eventually to identify putative mutations that gave rise to that bacteria’s stressor resistance.
The Bacterial Genomics program provides participants with the skills to conduct lab experiments and research that can tackle complex, real-world life science problems from a microscopic to a macroscopic level. Participants will also be mentored by genomic scientists and have the chance to meet with guest lecturers and speakers who are excited to share years of genomic research.
Key Dates and Deadlines
-
December 12th, 2024
Applications Open -
January 24th, 2025
Deadline for international applications -
February 21st, 2025
Deadline for domestic applications
-
Mid-April 2025
Admissions decisions released -
June 2025
Programs begin mid-late June
Is the Summer Science Program in Bacterial Genomics Right for You?
Applications are open each winter to current high school juniors who have completed or are in the process of completing any level of high school biology and algebra II by June for credit and a grade. We do NOT require AP or advanced-level classes. Self-study does not qualify.
Applicants must be at least 15 years old but not yet 19 during program operation. Current freshmen, sophomores, and seniors are not eligible.
Program Dates & Campuses
-
Albion College
June 8 – July 13 -
Pacific University
June 15 – July 20 -
Lehigh University
June 22- July 27 -
Purdue University
June 29 – August 3 -
University of Guelph
July 6 – August 10
2025 Research Project: Antibiotic Resistance and Directed Evolution
When any living thing reproduces, its DNA is copied – a process subject to random copying errors. Those copying errors are called “mutations,”” and they can be detrimental, neutral, or beneficial – and interestingly, a beneficial mutation under certain conditions might be detrimental in another! For instance, mutations that frequently occur to combat certain antibiotics happen in the protein machinery of the cell. These cells can survive the antibiotic but might grow overall slower than the ancestral strain. While antibiotics are a common stressor studied in the lab (and rightly so, as antibiotic resistance in bacteria is serious public health concern across the globe) these stressors can include innumerable things such as UV radiation, heavy metals, oxidative stress, and even too much or too little salt.
In the “Cell Stress and Directed Evolution” research project, you will study and stimulate the growth of a non-pathogenic bacteria while exposing it to a specific stressor to apply evolutionary pressure and select for stress-resistance mutations. This process is called “directed evolution” and is a key technique used by researchers to study both evolution and genetics. The identified mutation will help us better understand stressor resistance and gain insight into combatting public health problems worldwide.
A Preview of the Experience
You will work with a team of three to maintain the constant growth of a non-pathogenic bacteria under ever-increasing stress (and therefore selective pressure) using a custom chemostat, a device used to cultivate microorganisms. The chemostat includes components for mixing and aeration of the growth media, optical growth rate monitoring, and feedback control of stressor delivery to the culture. Teams will then sequence their evolved strains, re-create the full genome, and map any mutations that arose as a result of the incomplete growth suppression.
Summer Science Program faculty will teach the underlying science and hands-on techniques that teams will use in their research, including:
- Microbiology: directed evolution, aseptic technique, growth rate calculations, stress resistance characterization, natural transformation
- Molecular biology: genomic DNA extraction and quantification, PCR and PCR cleanup, agarose gel electrophoresis
- Instrumentation: micropipetters, chemostats, microplate readers, e-lab notebook software
- Bioinformatics: Galaxy (an open-access data analysis platform) for read trimming, genome assembly (de novo and from reference genomes) and quality control, gene annotation, SNP and mutation calling
- Academic reading, writing, and presenting: data visualization, journal article comprehension and summary, collaborative manuscript authorship, collaborative poster authorship and defense
Read Our Blog
Learn about the day-to-day experiences of the Summer Science Program in Bacterial Genomics, get to know notable Genomics alumni who now lead impactful careers, and stay updated on any major announcements through our blogs below.

NMSU Genomics – Day 25 (last one!): Vageesh’s Blog
July 31, 2024
Friday Today, I began my day walking back to Pinon at 12:30 AM after a long day of bioinformatics. As usual, Brett, Kano, and I walked to Foster for the morning lectures. We attended lectures from Ms. Alexa, Ms. Ceilidh, Mr. Shakil, and Mr. Joey. Today’s lectures were quite interesting from the line dancing in […]

NMSU Genomics – Day 24: Emily’s Blog
July 31, 2024
I woke up to a peaceful morning. Awakening to the blaring eagle screeching that I call my alarm, I walked into the echoes of hysterical laughter of my suitemates, Angelina, Ritisha, and Chloe. What were they laughing at? I stared at our toilet, overflowing with soapy water (I hope). Turns out, sleep deprived Angelina found […]

NMSU Genomics – Day 23: Angelina’s Blog
July 31, 2024
ᴺᴼᵂ ᴾᴸᴬᵞᴵᴺᴳ : my way by frank sinatra ᴠᴏʟᴜᴍᴇ : ▮▮▮▮▮▮▯▯▯ 0:57 ━━━•──────── 3:59 ⇄ ◃◃ ⅠⅠ ▹▹ ↻ “It’s time to go to Carlsbad!” is what I wished I would’ve thought when I woke up this morning. Like an SNP being the difference between antibiotic resistant bacteria and non-resistant bacteria, one might […]











